2014 Innovators Awards | Profile
Building DNA in the Cloud
Penn State researcher Howard Salis created a simple tool for a complex process — DNA sequencing — and turned it into a highly scalable, on-demand system that serves scientists all over the world.
|
Category: IT Infrastructure and Systems
Institution: Penn State University
Project: DNA Compiler
Project lead: Howard Salis, assistant professor of biological and chemical engineering
Tech vendor/partner: Amazon Web Services
|
Howard Salis' Twitter bio sums up his work well: "Creating synthetic microbes from the bottom-up."
As assistant professor of biological and chemical engineering and synthetic biology at Penn State University, Salis develops physical models that predict how DNA is interpreted inside an organism. Specifically, the models predict the rates at which that DNA will cause the organism to produce the corresponding amount of protein. "We can use these models to rationally engineer organisms to carry out new activities including the production of biofuels, plastics and drugs," he said.

Howard Salis |
Several years ago, when Salis was a postdoctoral fellow at the University of California, San Francisco, he and other researchers would combine many different genetic parts, trying to engineer a genetic system to have a particular desired behavior. "There were so many possible combinations that we could have put together and yet we only had the time and resources to think of a few and see if they worked," he explained. "So a lot of the research was trial and error in that regard. But today using physical models we can actually calculate the thermodynamic properties of these different genetic parts and we can make predictions about how they will work together when put together. It is like AutoCAD for biology."
In 2009 he observed that his field of synthetic biology needed improved computer-aided design software for researchers to do their work more efficiently. In response, Salis, who said he has been programming since he was 12 years old, created and launched the DNA Compiler Web portal in early 2010.
In developing the DNA Compiler, he recognized that a streamlined user interface was important. "The calculations are complicated, but nobody will use it unless the interface is friendly, so we basically took a very complicated model and put a simple input/output relationship on top of it on a clearly designed Web site," Salis said. If you are a biologist, you don't need to know how the model works, he noted. You can copy and paste in your DNA sequences and get predictions back. You can also tell the algorithm what you would like to accomplish in terms of how much protein it should express, and then the algorithm will design for you a completely new DNA sequence that will achieve that outcome.
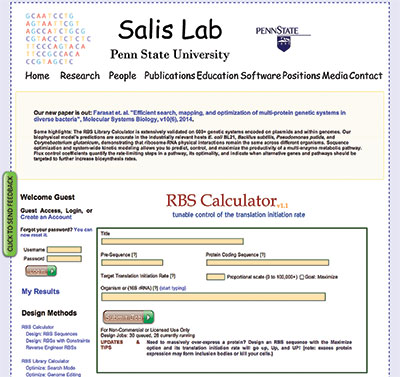
Behind the DNA Compiler Web portal's simple user interface, a powerful tool provides complicated calculations for genetic research. |
Within six months of Salis making the DNA Compiler available, researchers from Japan, China, the U.K, France, Finland and Sweden began using it. Salis created a Google Analytics map of usage. "Basically all the people who do synthetic biology research or metabolic engineering research at the institutions well known for those areas of research are using it," he said.
But the portal's popularity meant long queues on the server located in Salis' office. The server could run 16 simultaneous jobs and even then, there were 80 to 90 jobs in the queue. With the amount of data involved, jobs could take a significant amount of time and slow research. For example, more than 100 compute hours are required to predict the E. coli genome's protein production rates.
Although Penn State has its own high-performance computing resources, those systems are not connected to the Internet. "They are very finicky about people connecting to their computers from outside the network, which is understandable," Salis said.
The solution? Salis moved the DNA Compiler to the cloud using Amazon Web Services (AWS).
The portal now combines AWS Elastic Compute Cloud's AutoScaling groups for compute resources with Simple Queue Service (SQS) to decouple application components so they can be run independently, as well as Simple Storage Service for storage. This design has eliminated the need for researchers to wait in line for their jobs to be run — and it has made calculation times faster as well.
"We now have a nice on-demand computing system, where users from around the world can submit their jobs," Salis explained. "Compute nodes dynamically turn on in response to those jobs; they run them and then they turn themselves off."
Salis said there are certainly other cloud service providers to consider, but noted that Amazon has the largest compute cloud available. "Something like 40 percent of Internet traffic is Netflix, and it runs on Amazon AWS. I have my research lab, and people use my Web site; if Amazon were to have a server malfunction, they are not going to care about me. But they are going to care about Netflix. So it will get fixed really quickly. That is what you are signing up for: always-on access, almost scalable to infinity and low cost because of the economies of scale. Also, Amazon had already fully developed their platform by the time I started to use it, so I didn't have to learn how to use it while they were still developing it — which was not the case for other providers."
More than 2,000 biotechnology researchers designing over 30,000 synthetic DNA sequences have used the DNA Profiler over the past two years. The vision for this project is the global optimization of every nucleotide within a genome to perform a specific and useful task.
According to Salis, a crucial point is that cloud solutions such as AWS allow you to develop highly scalable on-demand resources that are connected to the Web. "So if there are some applications or interfaces that someone would like to develop that have to be connected very broadly to the Internet, it is much better to use a computing cloud environment than a dedicated hardware environment."
For a person running a research lab, if there is a problem that requires some intensive computing but you only need to solve it once, you may not want to deal with the hassle of buying or using institutional computational resources. "But if you have access to the compute cloud," Salis explained, "you can solve that problem in a short period of time using the exact same software you would normally use."
|
For more information on the Campus Technology Innovators program, visit the awards site.
|

